Specificity, length and luck drive gene rankings in association studies
·Projecting cells into different spaces based on experimental conditions
This time we are looking into this new article, Spence et al. 2025.
The aim of the study is to find out the differences between GWAS significant SNVs and burden tests identified top LOF variants. What researchers typically found that given the same phenotype, the LOF variants identified in burden tests do not rank top in GWAS; and the GWAS identified top hits do not have clear phenotypic effects on their associated genes.
In the study, the authors analysed the GWAS and LoF burden tests for 209 quantitative traits in the UK Biobank. The key finding of the article is that 1) burden tests prioritize trait-specific genes but with a small effect; 2) GWAS focuses on SNPS that affect pleiotropic genes in specific tissues. The 2 tests are complementary and highlight genes with different attributes to the phenotype.
Trait importance vs trait specificity
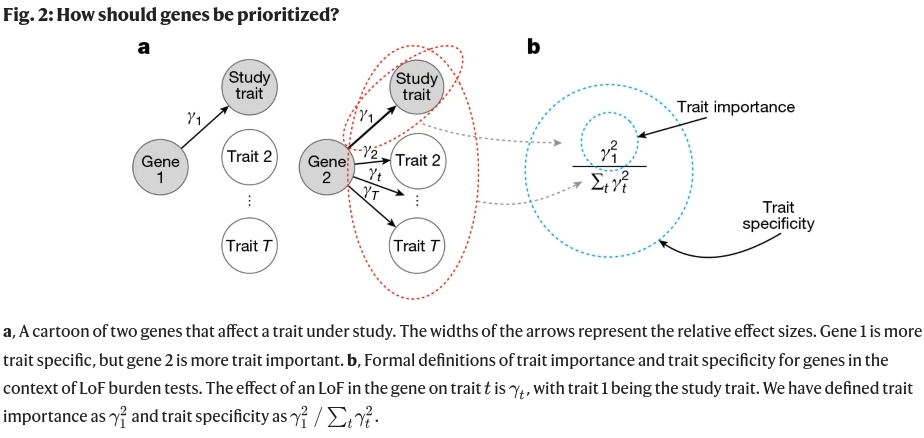
In the article the authors defined at,/sub> as the variant effect on a trait t, and Yt,/sub> as the LoF effect size of a gene. Trait importance is defined as its effect2 on the trait of interest. Trait specificity is defined is the trait importance relative to the importance across all relevant traits.
Lof Burden test prioritizes on trait specificity on gene level
The frequency of LoF variants can be approximated to Y2PLoF(1-PLoF), since the LoF can only occur in heterozygous.
LoF variants are highly dependent on the selection in heterozygotes.
A LoF variant on a gene with large effect, selection strength make the LoF becomes rare (PLoF small), and hence large standard errors on its frequency estimation and a small estimate on its strength of association (Z2:= (Y/SE(Y))2.
The authors found a delineated relationship between LoF variant rank vs trait importance, rather the burden test signals becomes stronger with trait specificity that was approximated as specfic expression of a gene.
GWAS prioritizes on trait specificity on variant level
In GWAS the strength of association is approximated as the variant effect/ the sum of all variant effects on a trait, and the metric is heritability.
Th authors found that variants acting on specifically expressed genes are prioritized higher by GWAS. The authors demonstrated the idea with 3 example on Fig 4a.

Variant 1 is a non-coding, with high trait specificity but its associated gene have low specificity to the trait. So it acts on a gene with pleiotropic effect but due to specific expression pattern, thus specific to a trait.
Variant 2 is coding, so it is very specific to a gene. But the gene is pleiotropic (phiG low), thus leading to a low trait specificity.
Variant 3 is coding, so it is again specific to a gene and the gene is very specific to the trait (phiG high). So it has a high GWAS score.
The more detailed explanation is that a LoF variant render a gene inactivation. The size of such effect depends on the gene effect and selection. If the gene only expressed in a small subset of tissue (i.e. highly specific expression), the overall gene effect is small and thus the selective pressure is relative lower (hets low). This leads to a higher variant frequency, and thus lower SE on its estimate and larger z.
GWAS, on the other hand, measure z with the variant effect on a gene combined with the gene effect on a trait.
The GWAS score is high either the variant control a pleiotropic gene (large gene effect) for specific expression; or the variant control a specifically expressed gene.
The authors found that if the variant is highly specific to the gene, and the gene has a solo effect to a trait (aka highly specific), this variant will be ranked high in GWAS.
Taken together, LoF burden test highlights genes with small effects and GWAS can identify variants act on large effect gene (high trait importance) as well.
determinants of the rank
The authors found that LoF burden test give stronger signal to long genes, which happens to be more likely to accumulate mutations and have more potential LoF sites.
On the contrary, GWAS results can be greatly affected by genetic drift, and variants could be identified as top hits in multiple traits.
Conclusion
The authors concluded by proposing to change the ranking methods to prioritize trait importance, which is very difficult to measure with real life data. “For example, trait-specific genes may be better drug targets due to reduced side effects, perhaps explaining why LoF burden evidence is more predictive of drug trial success than GWAS evidence. Yet, if pleiotropic genes can be targeted in a context-specific way, targeting the most trait-important genes may be more clinically impactful. “
This article gave a lot of thoughts on how we can analyse SNVs and what we are likely to derived from GWAS and LoF burden test. It also present the difficulties to deconstruct the phenotypic effect of a trait to specificity and importance ; and in both gene and variant levels.
Definitely a useful read, especially the supplementary information with detailed description on the GWAS and LoF burden test mathematical models and assumption.