MAGeCK - robust ranking for impacts of CRISPR screens
·Analysing CRISPR screens with MAGeCK.
In the beginning, I was reading about scMAGeCK, which is use for diciphering gene networks affected by CRISPR inhibition/aactivation from sgRNA read counts and scRNA-seq data.
When it is obvious that there are too much background information missing, I have to first sort out what MAGeCK does and this is what I talk about in this post.
MAGeCK (Li et al (2015)) was developed for finding out essntial genes in pooled CRISPR knockout screens. The typical setting of experiment that suitable for MAGeCK analysis are sequencing of cells transfected with pooled CRISPR library before and after a selection process. The sgRNA were identified and count from the sequencing data to produce a count matrix. Assuming that the importance of the targeted gens are implicated in the sgRNA abundance changes, MAGeCK estimates the collective effects of multiple sgRNAs targeting the same genes (normalising for sgRNA activities) and rank the importance of the targeted gene in a given condition.
More specifically, the counts of each sgRNA from multiple replicates were normalised by a size factor (s) that capture the technical variations of all sgRNAs (denoted i) in an experiment j, where $\hat{x}$ is the geometric mean of individual sgRNAi across experiments.
$ s_{j} = median \frac{x_{ij}}{\hat{x}} $
The abundances of the sgRNAs are then modeled in a NB distribution (means and variances of sgRNAs across experiments.) and the counts of a sgRNAi is updated.
MAGeCK calculate the mean and variances of sgRNAs in control condition; then compute the probability of the corrected count of the sgRNAi in treatment condition based on the Negative Binomial distribution modeled in control condition ($ NB(\mu_{control}, \sigma^2_{control}) $). Then the sgRNAs are ranked according to the NB p-values.
Robust rank aggregation (RRA) is employed to evaluate the importance of a targeted gene. The sgRNA rank is then converted to a percentile rank. Then, the probability (p(rk})) of obtaining k significant sgRNAs among N total sgRNAs in the pool in a null model threshold (5% significant level) is expressed as the binomial probability of picking such percentile rank in the null model.
To pool the multiple sgRNAs ranks targeting the same gene, MAGeCK only take the minimal p(rk) among the filtered sgRNAs (i.e. NB p-values $\le$ 0.05) as the representative $\rho$-value for the gene. The final p-value of the gene significance were obtained from permutation test that pair sgRNAs and genes randomly.
MAGeCK is sl applicable for RNAi screens.
Running MAGeCK
conda create -c bioconda -n mageckenv mageck
# To activate this environment, use
#
# $ conda activate mageckenv
#
# To deactivate an active environment, use
#
# $ conda deactivate
MAGeCK can be easily installed via bioconda.
Lets try a few of the tutorials and see how to perform MAGeck and us the downstream MAGeCKFlute (https://www.bioconductor.org/packages/devel/bioc/vignettes/MAGeCKFlute/inst/doc/MAGeCKFlute.html) to analyse the MAGeCK output.
Demo3 went through how to use MAGeCK to screen for essential genes in mouse ESC cells. We need the negative control (plasmid library ERR376998) and the treatment (sgRNA recovered from transfected ESC ERR37699).
The sgRNA template file is already prepared and available in the MAGeCK libraries yusa_library.csv.
mageck count -l yusa_library.csv -n escneg --trim-5 23 --sample-label "plasmid,ESC1" --fastq ERR376998.fastq ERR376999.fastq
First we run MAGeCK to count the sgRNAs in the fastq file. Here we trim away the first 23 bases from the fastq to remove the adapter, and label the sample as plasmid and ESC1.
Examining the “escneg.countsummary.txt” showed that upon ESC transfection, sgRNAs scored zero counts rose from 210 to 4590. And the Gini Index increased from 0.117 to 0.2014, showing that the counts of sgRNAs had become somewhat uneven.
The “escneg.count.txt” listed all the sgRNAs and their raw read counts in the two fastq files and “escneg.count_normalized.txt” stored the normalised counts.
mageck test -k escneg.count.txt -t ESC1 -c plasmid -n esccp
MAGeCK test compares the sgRNA difference between control (-c plasmid) and treatment (-t ESC1). “esccp.sgrna_summary.txt” listed out all the RRA analysis results of each sgRNA, the log2 fold-change and whether the sgRNA is significant. And “esccp.gene_summary.txt” listed all the significant genes, in this case essential genes for mouse ESC cells. The “gene_summary.txt” file is also the input for MAGeCKFlute.
MAGeCKFlute
library(MAGeCKFlute)
library(ggplot2)
file1 = file.path("/home/achu/Downloads/mageck-0.5.9.4/demo/demo3/esccp.gene_summary.txt")
# Read and visualize the file format
gdata = read.delim(file1, check.names = FALSE)
head(gdata)
gdata = ReadRRA(file1)
head(gdata)
# Read in sgRNA file
file2 = file.path("/home/achu/Downloads/mageck-0.5.9.4/demo/demo3/esccp.sgrna_summary.txt")
sdata = read.delim(file2)
head(sdata)
# Run the FluteRRA pipeline
FluteRRA(file1, file2, proj="esccp_Test", organism="mmu",
scale_cutoff = 1, outdir = "/home/achu/Downloads/mageck-0.5.9.4/demo/demo3/")
MAGeCKFlute (Wang et al (2019))has performed important genes identification, KEGG, Reactome, GO biological processes and protein complex enrichment analyses.
For example, MAGeCKFlute identified positive and negative selected genes in this screen.
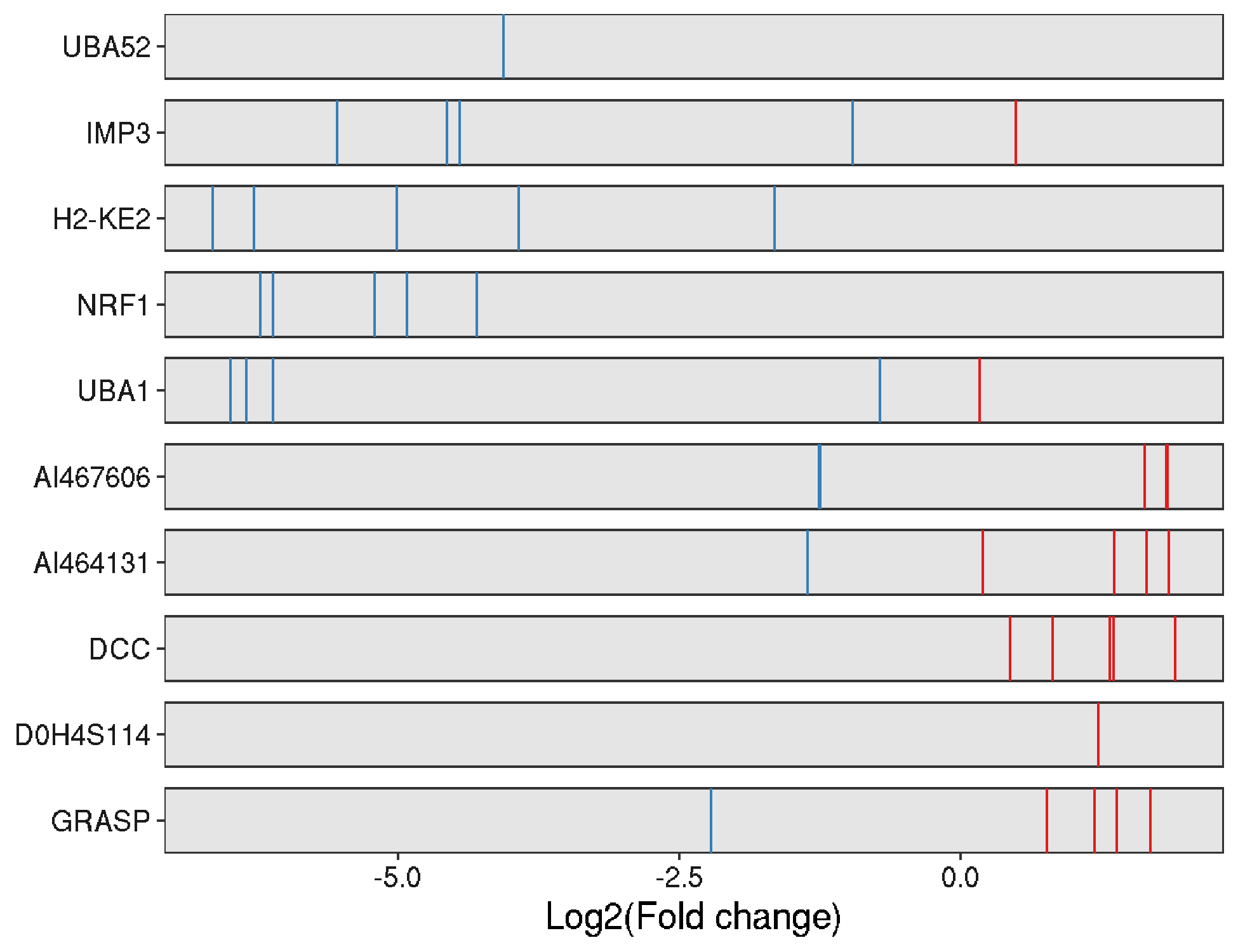
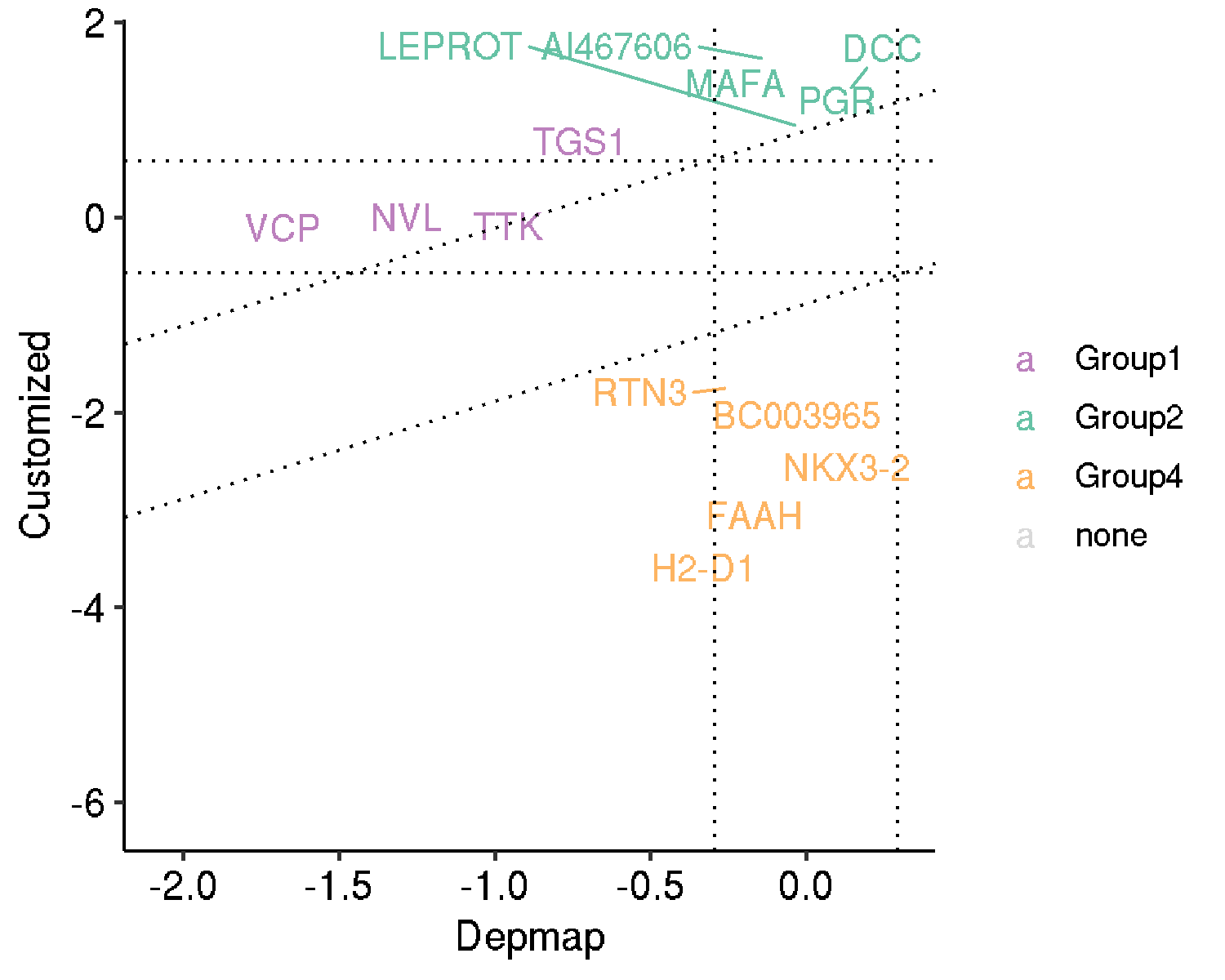
In addition, MAGeCKFlute computed the selection scores and the DepMap score. It showed that three groups of genes were identified and Group2 seems to be genes that had showed both positive score and positive DepMap score (thus positively selected in the ESC cells while important for cancer growth?).
Conclusion
So far, MAGeCK is very easy to use and has done a great job in identifying important genes and sgRNAs in a screen. I am a bit confused with the FluteRRA output; more explanations would be welcome. It would be good to read more about how its single-cell application is like, especially for screens with multiplexed sgRNAs.