Revisiting CHIP-seq
·How far have we progressed from the noises?
So instead of catching up with the single-cell stuff, I am looking into CHIP-seq this time. It is because I was given a small project to compare two CHIP-seq datasets (Law 2010 vs Sarma 2014). The two studies used the same ATRX antibodies to interrogate the silencing mechanism of the protein and arrived at somewhat different conclusions. My takes from the papers are that they used the CHIP-seq experiments to address very different questions to begin with, so it is nothing surprising that they report slightly different functional role of ATRX. Nonetheless, it would be interesting to compare the two CHIP-seq datasets to see if they agreed with each other or not.
A small comment is that using ascp makes life a lot better; it makes downloading the SRA files ways faster with a set bandwidth and more stable. So byebye SRAtools kits.
The first step after sequencing reads preprocessing is alignment. I used botwie2 to retrieve up to 50 multiple-mapped reads for the datasets. There were discussions that using more than 50 multiple mapped reads do not make more information gain, so I set the cap as 50 to reduce some computing time. There are new literature coming up to utilise these reads (i.e.ShortStackV3) to increase sensitivity of peak detection. Why shall we rescue these reads? Because it made up of 20 to 50% of the data! And assigning them to their likely origin rather than leaving them as randomly assigned is crucial for peak detection, which is assessing the reads enrichment in a small region of the genome. Still, I think it would be important to try a few more alignment methods and fine-tune the alignment parameters as good alignments makes detection way more accurate but not the other way round.
Regardless, the next thing is to try MACS2 in peak detection and finally retrieve the shared peaks between replicates using IDR. I have set the effective genome size (-g) as 1.87e9 for mm10, as suggested by the MACS2 manual. But it maybe better to consult deepTools2 for a more precise guess as I have used the unmasked genome.
macs2 callpeak -t ${rddir}/SRR1258446_sort.bam \
-c ${rddir}/SRR1258447_sort.bam \
-s 0 \
--slocal=1000 \
-f BAM -g 1.87e9 \
-n 2014_ATRX_rep1 \
--outdir ${rddir}/macs2
Of course, looking into IGV to have a feel of how the peaks look like woule to useful too. I would like to use DeepTools2 to QC the samples. Well that’s the plan if I can install it without root access…
Understanding the MACS2 output
The focus is on the Narrowpeak file, informing us where the peaks and summits are with a peak score indicative of how convening the peak is. There are a lot of peaks detected, so I moved on the IDR to filter for high-confidence peaks to have a first glimpse of how the data looks like.
The first impression from the IDR plot is confusing! The Day0 and Day7 ES cells dataset generated from Sarma et. al. 2014 had high alignment rate but very low sensitivity. Very few peaks were detected and the DNA input control almost shared the same read distribution patterns as the CHIP-seq data (with lower read-depth). IDR analyses reported very low reproducibility and The Day7 dataset failed IDR analysis due to insufficient number of peaks. Such inconsistency could be resulted from the instability of chromatin in ES cells, well they are still under a lot of reprogramming at this stage. The seemingly spurious results may be also due to the high read-depth in the negative controls, but what caused this exactly in the experiment?
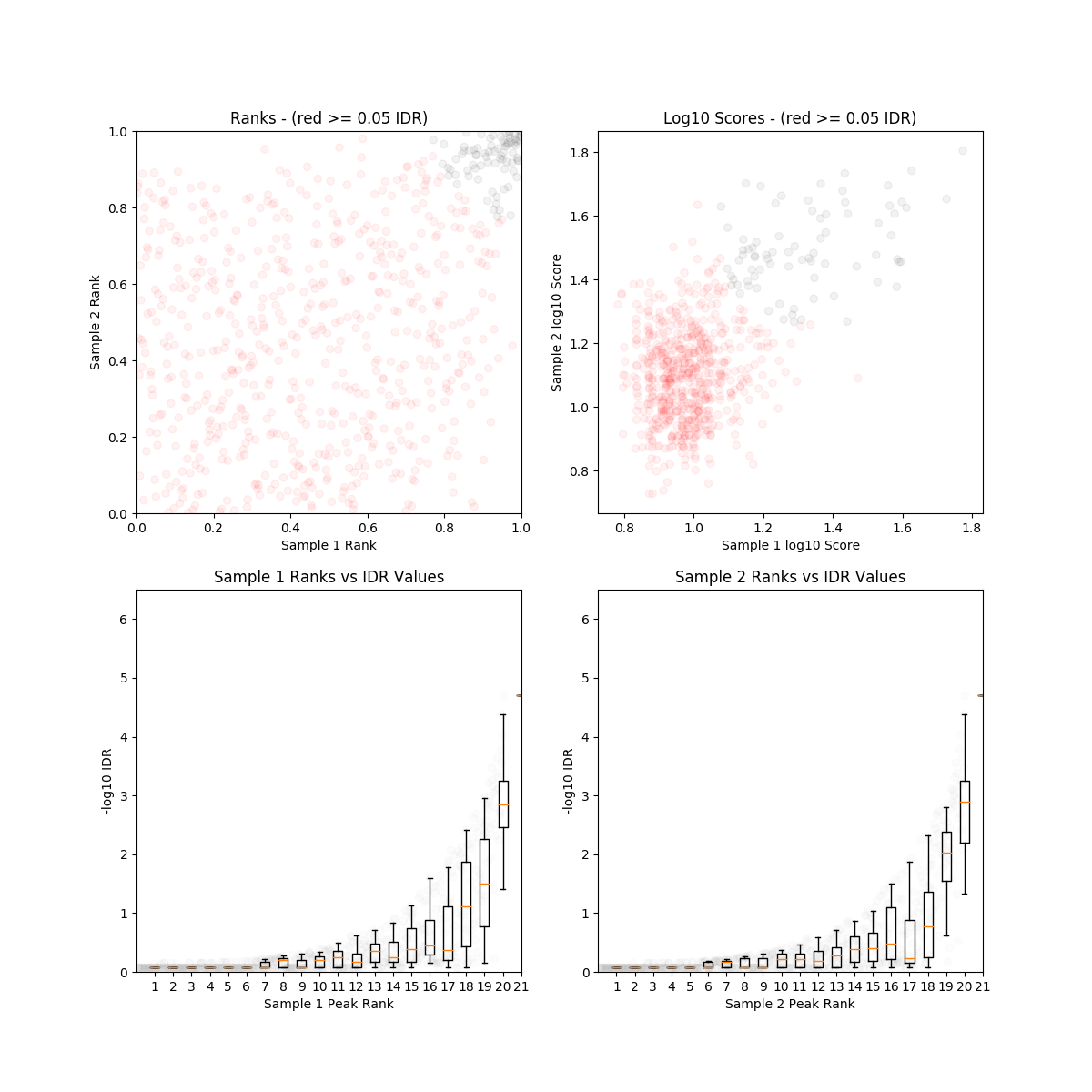
On another hand, the law 2010 et. al. data has very high reproducibility between replicates and there were a lot of peaks. The CHIP-seq was also done on mouse ES cells. Is such consistency observed due to high experimental quality or the ES cell line used in this study had more established chromatin status?
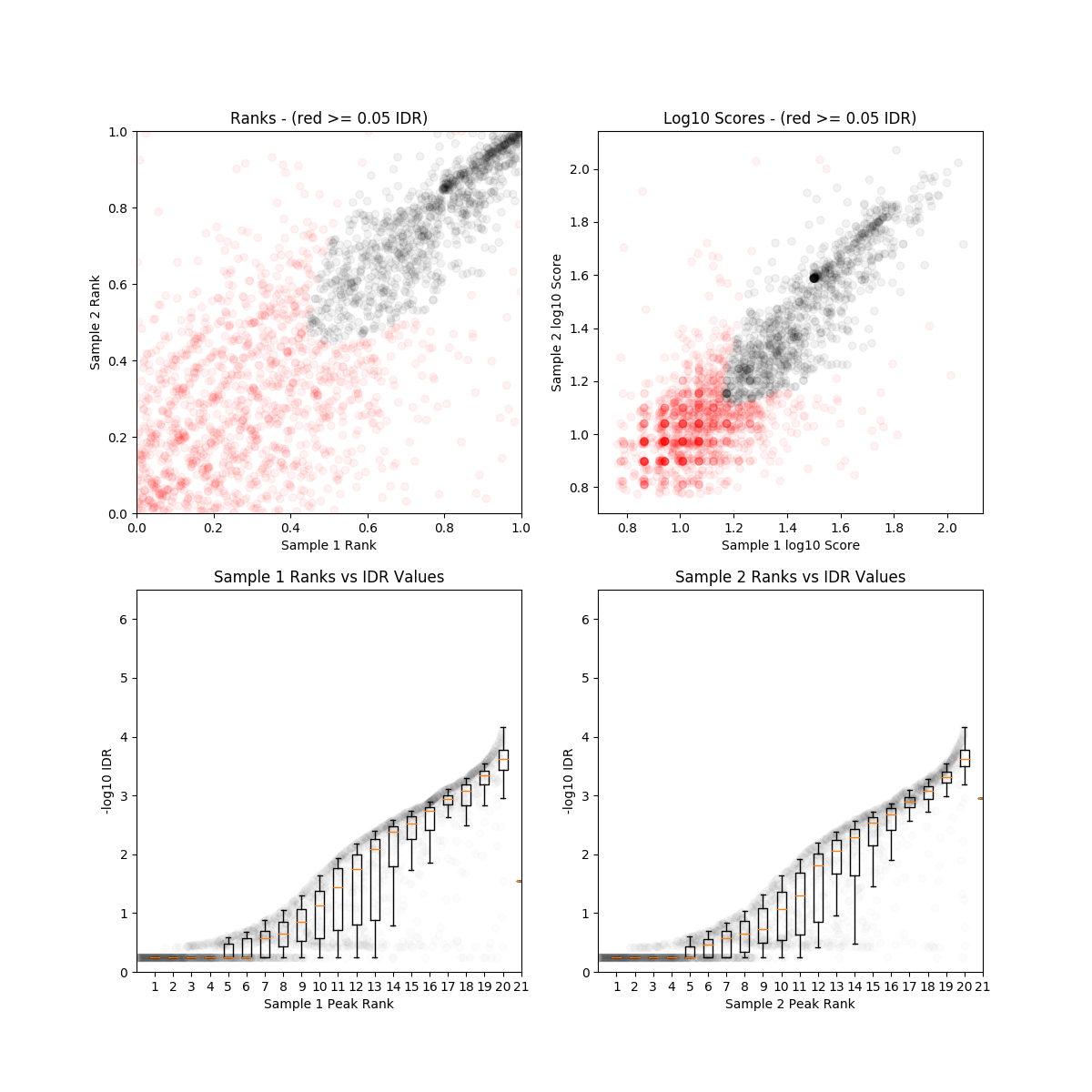
The differences between this two datasets pose an eminent challenge of reusing published data. It is fantastic news that we are now sharing our empirical data but also a nightmare that we rarely put efforts in standardization of how we collect the data. In the end of the day, if we cannot utilize and build upon this data for further scientific discoveries, what a waste?